Earlier this month the UK Cereal Pathogen Virulence Survey (UKCPVS) held its annual stakeholder meeting. Although there were no major surprises, CPM finds out there’s more going on than meets the eye.
I’d hesitate to describe the yellow rust population as stable.
By Lucy de la Pasture
For the first time in several years the UKCPVS was able to report that last year all appeared calm, with no particular reason to expect varieties to behave differently to their resistance ratings on the AHDB Recommended List.
Yet underneath the headline, the field of science studying pathogen virulence is paddling furiously to keep up and maybe even get ahead of the constant changes in the UK’s pathogen population, particularly for yellow rust.
“I’d hesitate to describe the yellow rust population as stable,” says Sarah Holdgate, plant pathologist at NIAB who leads the UKCPVS project. “But what I can say is that there have been no major shifts since the last big change in 2016, when the Red 24 group of isolates came to prominence. But in 2018 we have seen some stabilisation, with a couple of pathotypes most dominant in the samples tested.”

Sarah Holdgate says their were no step-changes in pathogen populations last year.
The Red group is related to the incursion of the original Warrior group of yellow rust in 2011, now renamed the Pink group. Since the original incursion, subtle changes in virulence patterns has occasionally led to some varieties unexpectedly breaking down badly to yellow rust infection.
Since 2011, a total of 17 wheat varieties have dropped in resistance rating for this reason, with seven varieties being down-rated by two RL rating points or more after the 2016 season, when the Red 24 group of pathotypes was identified as the prime cause of unexpected behaviour in the field.
Guide to new yellow rust groups

In 2017, UKCPVS reported that five new pathotypes of yellow rust had been identified within the survey, with Red 30 and a further complex pathotype (unassigned to any group) giving particular cause for concern. These two pathotypes, together with Red 24 (most common), Red 28 and Red 26 were put into adult and seedling trials which demonstrated all varieties were susceptible to one or more of the isolates except Costello, KWS Crispin, KWS Siskin, KWS Trinity and KWS Firefly, reports NIAB’s Amelia Hubbard.
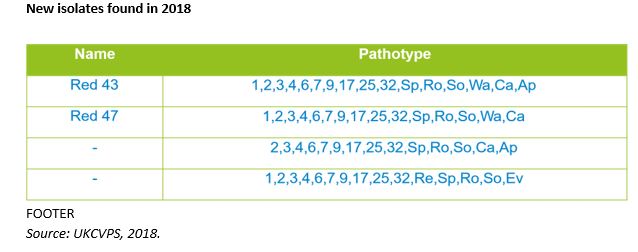
“In 2018 we identified some new pathotypes in the yellow rust isolates tested. Red 27 was the most common pathotype in 2018 and was first seen the year before in Graham and Shabras (rated 8 on RL),” she explains.
“Yellow rust found on variety Freiston (9) was identified as a new pathotype to the UK, Red 43 and we found Pink 10 on Revelation (9). Also of interest was yellow rust found on KWS Siskin (9), but unfortunately the sample failed during incubation so we weren’t able to test for its pathotype. This normally means that it isn’t a true infection and KWS Siskin is still likely to be resistant.”
The genetic data for 2018 wasn’t available at the time of the UKCPVS meeting to give a complete picture of any population changes, but Amelia was able to confirm the most commonly found isolates were Red 27 and Red 3 based on pathology data produced at NIAB.
“Of the new isolates found in 2018, Red 43 and Red 47 may be associated with slightly higher than expected levels of disease, but this needs to be confirmed,” she adds.
Common isolates found in 2018

The field pathogenomics utilised within the UKCPVS was developed as a partnership between NIAB, the Earlham Institute and John Innes Centre. Aside from surveillance, field pathogenomics can be used to answer fundamental questions about how the yellow rust develops over time and space.
“The yellow rust population is very diverse and changes can happen extremely quickly,” explains plant pathologist, Chris Judge. How changes occur within the field throughout the season at the pathotype level is something he set out to try and understand in a two-year series of trials at sites across the UK.
Getting a handle on whether isolates found in the autumn are the same ones responsible for causing epidemics later in the season, or whether isolates found in one part of the field are the same as those found a few metres away, could possibly help us provide an early warning of changes in the yellow rust population, he comments.
Chris intensively scouted the trial fields for yellow rust infections, marking their location within fields and sending samples for RNA sequencing to elucidate their genotype.
“In the 2015-16 season we had trials at 14 different sites, and we found pathotypes mostly from the Red group, with few from the Pink and Purple groups. The following season all the pathotypes were found to be from the Red group and there was much more diversity than we had expected,” he explains.
“The intensive monitoring found a total of 36 different pathotypes, with some not seen within the UKCPVS programme of testing. Six of the most commonly found pathotypes were found in every region of the country,” adds Chris.
One of the interesting findings of the study was the diversity found at yellow rust infection loci. “We found diversity in each affected ring of plants, which highlights that although yellow rust may be in the same area within the field, it may not be the same pathotype.
“As well as variation in space, we found the same is true in time. Testing of isolates found in Jan found they were different to the pathotypes found in March at the same location within the field. This means early season pathotypes may be lost later in the season and different pathotypes gained.
“This means it’s impossible to predict how yellow rust may behave later in the season from sequencing the infections found earlier in the season,” he comments.
Why yellow rust exhibits so much variation in time has two possible explanations, says Chris. “It may be because of simple variation between samples, but I believe it’s more likely that new pathotypes emerge when new spores blow into the crop.”
On a practical level, the study has shown up some potential holes in the sampling process when agronomists collect yellow rust samples for UKCPVS testing.
“In light of the study, we suggest sending in 4-5 leaves from a 2-4m2 area. When samples arrive at NIAB, we sample a single lesion for spores from a single pustule, so wrapping leaves separately will mean we have a better chance of picking up the pathotypes that may be present in an infection loci,” adds Sarah.
The mind-boggling world of molecular genetics may also be able to provide information to plant breeders which will be ultimately be of use to aid disease management, explains Dr Diane Saunders of John Innes Centre.
Field pathogenomics isn’t only useful to help understand changes in the pathogen population, it can also give a valuable insight into the other side of the coin – how the host plant interacts with the pathogen, she says.
“Identifying which pathways of the host cell are targeted by the pathogen is crucial to get a better understanding of the mechanisms involved in disease response. By looking at the interaction of the pathogen and host we can work out genes which are being turned on or off,” she explains.
JIC PhD student, Pilar Corredor-Moreno, has been researching just this – using the varieties Solstice, Cordiale, Oakley and Santiago. The latter two varieties are from closely related genetic lineages, otherwise the varieties are genetically distantly related to one another and show differing levels of susceptibility to yellow rust.
“Pilar has found a difference in the expression of genes related to the chloroplasts of susceptible varieties and those with greater resistance to yellow rust. She’s identified one gene – encoding a branched-chain amino acid aminotransferase – which appears involved in the plant’s defence response. By knocking out the function of the gene, it’s possible to elucidate its role in disease progression.
“This information can be used by breeders in their breeding programmes and although the resistance trait for this gene may have an associated yield penalty, this approach is starting to uncover new knowledge into how pathogens manipulate their host that could ultimately help breeders to produce new varieties using this insight that have an acceptable yield potential,” comments Diane.
Revised timing advice on T2 for ramularia
Syngenta trials suggest chlorothalonil applied at the T2 riming for ramularia control should be used slightly earlier than the conventional GS45-59 timing.
According to their field technical manager Iain Hamilton, Ramularia collo-cygni, traditionally a problem in Scotland, has become more widespread over the past five years, with infections occurring from the north to the south of the UK.
Infection starts from the seed, he says, though can also come from crop debris, with the disease then entering asymptomatic phase. The damaging phase is triggered later in the season when the crop is put under stress, causing the pathogen to produce a toxin which results in spotting symptoms on leaves.
“With no effective seed treatments against ramularia, and variety resistance ratings not currently available, the multi-site fungicide Bravo (chlorothalonil) has become a mainstay for tackling it, especially since the reduced performance or resistance seen with other fungicide groups.
“Historically, Bravo has been applied as part of the conventional T2 fungicide spray at the awns emerging to ears emerged timing (GS49-59). But new results from last season showed a much greater ramularia reduction was achieved on the top three leaves by spraying earlier than this – either at the flag leaf emerged timing (GS39) or at booting (GS45),” he explains.
T2 applications at the latest application timing, GS59 showed it was too late and control of ramularia had already been lost.
“These results were echoed by improvements in green leaf area by spraying at these earlier timings and, more importantly, by increases in yield.”
Based on the new findings, Iain suggests growers should consider these earlier spray timings in winter and spring barley – either by applying Bravo at GS39 followed by an SDHI, such as Elatus Era (benzovindiflupyr+ prothioconazole) as ears emerge at GS55-59; or by applying a tank mixture of Bravo and Elatus Era at GS39-45. The former approach may suit situations where late brown rust is expected, he notes.
“Bringing the Bravo spray forward from the traditional T2 timing fits with what we know about ramularia. Although this is just one year’s results, chlorothalonil is a protectant fungicide so needs to be used preventatively, so it’s important to treat the crop before symptoms appear.
“Applying Bravo at GS39 isn’t necessarily an extra spraying operation. An ethephon-based plant growth regulator (Terpal) is often sprayed on barley at this time anyway. Other trials have also shown better net blotch control, better green leaf retention, better yield and better margin over fungicide cost with the GS39-45 Bravo plus Elatus Era timing.”
Iain estimates that in general, there’s a 14-day gap between GS39 and GS49 which makes a fungicide sequence at these timings possible. “But for spring barley it’s unlikely to be a practical option as the crop can race through its growth stages in just a few days.”




